Part 3: Use an nf-core module¶
In this third part of the Hello nf-core training course, we show you how to find, install, and use an existing nf-core module in your pipeline.
One of the great benefits of working with nf-core is the ability to leverage pre-built, tested modules from the nf-core/modules repository. Rather than writing every process from scratch, you can install and use community-maintained modules that follow best practices.
To demonstrate how this works, we'll replace the custom collectGreetings module with the cat/cat module from nf-core/modules in the core-hello pipeline.
Note
This part of the course assumes you have completed Part 2: Rewrite Hello for nf-core and have a working core-hello pipeline.
If you did not complete Part 2 or want to start fresh for this part, you can use the core-hello-part2 solution as your starting point.
Run this command from within the hello-nf-core/ directory:
This gives you a fully functional nf-core pipeline ready for adding modules.
1. Find and install a suitable nf-core module¶
First, let's learn how to find an existing nf-core module and install it into our pipeline.
We'll aim to replace the collectGreetings process, which uses the Unix cat command to concatenate multiple greeting files into one.
Concatenating files is a very common operation, so it stands to reason that there might already be a module in nf-core designed for that purpose.
Let's dive in.
1.1. Browse available modules on the nf-core website¶
The nf-core project maintains a centralized catalog of modules at https://nf-co.re/modules.
Navigate to the modules page in your web browser and use the search bar to search for 'concatenate'.
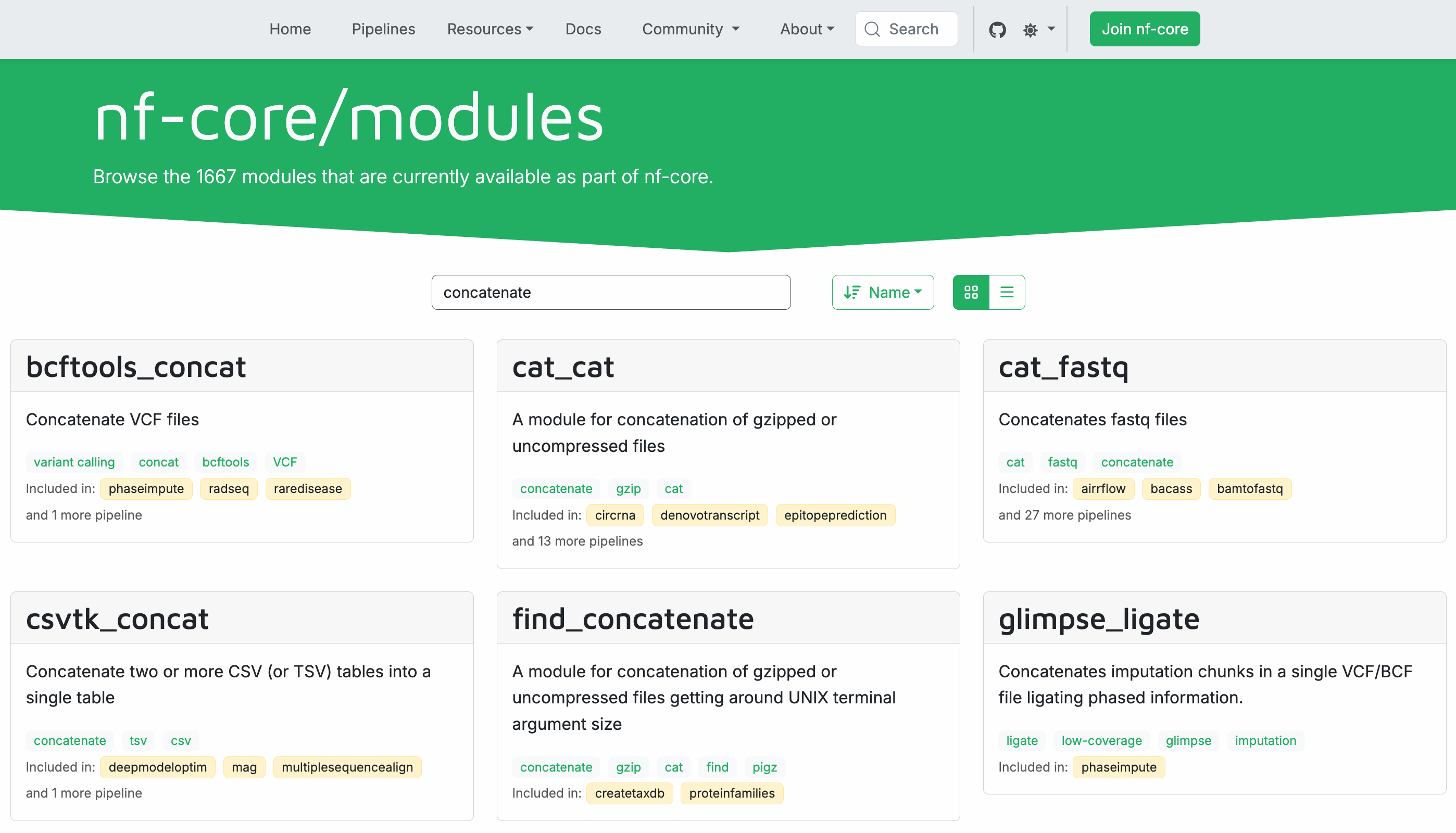
As you can see, there are quite a few results, many of them modules designed to concatenate very specific types of files.
Among them, you should see one called cat_cat that is general-purpose.
Module naming convention
The underscore (_) is used as a stand-in for the slash (/) character in module names.
nf-core modules follow the naming convention software/command when a tool provides multiple commands, like samtools/view (samtools package, view command) or gatk/haplotypecaller (GATK package, HaplotypeCaller command).
For tools that provide only one main command, modules use a single level like fastqc or multiqc.
Click on the cat_cat module box to view the module documentation.
The module page shows:
- A short description: "A module for concatenation of gzipped or uncompressed files"
- Installation command:
nf-core modules install cat/cat - Input and output channel structure
- Available parameters
1.2. List available modules from the command line¶
Alternatively, you can also search for modules directly from the command line using nf-core tools.
This will display a list of all available modules in the nf-core/modules repository, though it's a little less convenient if you don't already know the name of the module you're searching for.
However, if you do, you can pipe the list to grep to find specific modules:
Just keep in mind the that grep approach will only pull out results with the search term in their name, which would not work for cat_cat.
1.3. Get detailed information about the module¶
To see detailed information about a specific module from the command line, use the info command:
This displays documentation about the module, including its inputs, outputs, and basic usage information.
Output
,--./,-.
___ __ __ __ ___ /,-._.--~\
|\ | |__ __ / ` / \ |__) |__ } {
| \| | \__, \__/ | \ |___ \`-._,-`-,
`._,._,'
nf-core/tools version 3.4.1 - https://nf-co.re
╭─ Module: cat/cat ─────────────────────────────────────────────────╮
│ 🌐 Repository: https://github.com/nf-core/modules.git │
│ 🔧 Tools: cat │
│ 📖 Description: A module for concatenation of gzipped or │
│ uncompressed files │
╰────────────────────────────────────────────────────────────────────╯
╷ ╷
📥 Inputs │Description │Pattern
╺━━━━━━━━━━━━━━━━━┿━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┿━━━━━━━╸
input[0] │ │
╶─────────────────┼──────────────────────────────────────────┼───────╴
meta (map) │Groovy Map containing sample information │
│e.g. [ id:'test', single_end:false ] │
╶─────────────────┼──────────────────────────────────────────┼───────╴
files_in (file)│List of compressed / uncompressed files │ *
╵ ╵
╷ ╷
📥 Outputs │Description │ Pattern
╺━━━━━━━━━━━━━━━━━━━━━┿━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┿━━━━━━━━━━━━╸
file_out │ │
╶─────────────────────┼─────────────────────────────────┼────────────╴
meta (map) │Groovy Map containing sample │
│information │
╶─────────────────────┼─────────────────────────────────┼────────────╴
${prefix} (file) │Concatenated file. Will be │ ${file_out}
│gzipped if file_out ends with │
│".gz" │
╶─────────────────────┼─────────────────────────────────┼────────────╴
versions │ │
╶─────────────────────┼─────────────────────────────────┼────────────╴
versions.yml (file)│File containing software versions│versions.yml
╵ ╵
💻 Installation command: nf-core modules install cat/cat
This is the exact same information you can find on the website.
1.4. Install the cat/cat module¶
Now that we've found the module we want, we need to add it to our pipeline's source code.
The good news is that the nf-core project includes some tooling to make this part easy.
Specifically, the nf-core modules install command makes it possible to automate retrieving the code and making it available to your project in a single step.
Navigate to your pipeline directory and run the installation command:
The tool may first prompt you to specify a repository type. (If not, skip down to "Finally, the tool will proceed to install the module.")
Output
,--./,-.
___ __ __ __ ___ /,-._.--~\
|\ | |__ __ / ` / \ |__) |__ } {
| \| | \__, \__/ | \ |___ \`-._,-`-,
`._,._,'
nf-core/tools version 3.4.1 - https://nf-co.re
WARNING 'repository_type' not defined in .nf-core.yml
? Is this repository a pipeline or a modules repository? (Use arrow keys)
» Pipeline
Modules repository
If so, press enter to accept the default response (Pipeline) and continue.
The tool will then offer to amend the configuration of your project to avoid this prompt in the future.
Output
Might as well take advantage of this convenient tooling! Press enter to accept the default response (yes).
Finally, the tool will proceed to install the module.
Output
The command automatically:
- Downloads the module files to
modules/nf-core/cat/cat/ - Updates
modules.jsonto track the installed module - Provides you with the correct
includestatement to use in your workflow
Note
Always make sure your current working directory is the root of your pipeline project before running the module installation command.
Let's check that the module was installed correctly:
Directory contents
You can also verify the installation by asking the nf-core utility to list locally installed modules:
INFO Repository type: pipeline
INFO Modules installed in '.':
┏━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━┓
┃ Module Name ┃ Repository ┃ Version SHA ┃ Message ┃ Date ┃
┡━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━┩
│ cat/cat │ nf-core/modules │ 41dfa3f │ update meta.yml of all modules (#8747) │ 2025-07-07 │
└─────────────┴─────────────────┴─────────────┴────────────────────────────────────────┴────────────┘
This confirms that the cat/cat module is now part of your project's source code.
However, to actually use the new module, we need to import it into our pipeline.
1.5. Update the module imports¶
Let's replace the include statement for the collectGreetings module with the one for CAT_CAT in the imports section of the workflows/hello.nf workflow.
As a reminder, the module install tool gave us the exact statement to use:
include { CAT_CAT } from '../modules/nf-core/cat/cat/main'`
Note that the nf-core convention is to use uppercase for module names when importing them.
Open up core-hello/workflows/hello.nf and make the following substitution:
Notice how the path for the nf-core module differs from the local modules:
- nf-core module:
'../modules/nf-core/cat/cat/main'(referencesmain.nf) - Local module:
'../modules/local/collectGreetings.nf'(single file reference)
The module is now available to the workflow, so all we need to do is swap out the call to collectGreetings to use CAT_CAT. Right?
Not so fast.
At this point, you might be tempted to dive in and start editing code, but it's worth taking a moment to examine carefully what the new module expects and what it produces.
We're going to tackle that as a separate section because it involves a new mechanism we haven't covered yet: metadata maps.
Note
You can optionally delete the collectGreetings.nf file:
However, you might want to keep it as a reference for understanding the differences between local and nf-core modules.
Takeaway¶
You know how to find an nf-core module and make it available to your project.
What's next?¶
Assess what a new module requires and identify any important changes needed in order to integrate it into a pipeline.
2. Assess the requirements of the new module¶
Specifically, we need to examine the interface of the module, i.e. its input and output definitions, and compare it to the interface of the module we're seeking to replace. This will allow us to determine whether we can just treat the new module as a drop-in replacement or whether we'll need to adapt some of the wiring.
Ideally this is something you should do before you even install the module, but hey, better late than never.
(For what it's worth, there is an uninstall command to get rid of modules you decide you no longer want.)
Note
The CAT_CAT process includes some rather clever handling of different compression types, file extensions and so on that aren't strictly relevant to what we're trying to show you here, so we'll ignore most of it and focus only on the parts that are important.
2.1. Compare the two modules' interfaces¶
As a reminder, this is what the interface to our collectGreetings module looks like:
| modules/local/collectGreetings.nf (excerpt) | |
|---|---|
The collectGreetings module takes two inputs:
input_filescontains one or more input files to process;batch_nameis a value that we use to assign a run-specific name to the output file, which is a form of metadata.
Upon completion, collectGreetings outputs a single file path, emitted with the outfile tag.
In comparison, the cat/cat module's interface is more complex:
The CAT_CAT module takes a single input, but that input is a tuple containing two things:
metais a structure containing metadata, called a metamap;input_filescontains one or more input files to process, equivalent tocollectGreetings'sinput_files.
Upon completion, CAT_CAT delivers its outputs in two parts:
- Another tuple containing the metamap and the concatenated output file, emitted with the
file_outtag; - A
versions.ymlfile that captures information about the software version that was used, emitted with theversionstag.
Note also that by default, the output file will be named based on an identifier that is part of the metadata (code not shown here).
This may seem like a lot to keep track of just looking at the code, so here's a diagram to help you visualize how everything fits together.
You can see that the two modules have similar input requirements in terms of content (a set of input files plus some metadata) but very different expectations for how that content is packaged. Ignoring the versions file for now, their main output is equivalent too (a concatenated file), except CAT_CAT also emits the metamap in conjunction with the output file.
The packaging differences will be fairly easy to deal with, as you'll see in a little bit. However, to understand the metamap part, we need to introduce you to some additional context.
2.2. Understanding metamaps¶
We just told you that the CAT_CAT module expects a metadata map as part of its input tuple. Let's take a few minutes to take a closer look at what that is.
The metadata map, often referred to as metamap for short, is a Groovy-style map containing information about units of data. In the context of Nextflow pipelines, units of data can be anything you want: individual samples, batches of samples, or entire datasets.
By convention, an nf-core metamap is named meta and contains the required field id, which is used for naming outputs and tracking units of data.
For example, a typical metadata map might look like this:
Or in a case where the metadata is attached at the batch level:
Now let's put this in the context of the CAT_CAT process, which expects the input files to be packaged into a tuple with a metamap, and outputs the metamap as part of the output tuple as well.
| modules/nf-core/cat/cat/main.nf (excerpt) | |
|---|---|
As a result, every unit of data travels through the pipeline with the relevant metadata attached. Subsequent processes can then readily access that metadata too.
Remember how we told you that the file output by CAT_CAT will be named based on an identifier that is part of the metadata?
This is the relevant code:
| modules/nf-core/cat/cat/main.nf (excerpt) | |
|---|---|
This translates roughly as follows: if a prefix is provided via the external task parameter system (task.ext), use that to name the output file; otherwise create one using ${meta.id}, which corresponds to the id field in the metamap.
You can imagine the input channel coming into this module with contents like this:
ch_input = [[[id: 'batch1', date: '25.10.01'], ['file1A.txt', 'file1B.txt']],
[[id: 'batch2', date: '25.10.26'], ['file2A.txt', 'file2B.txt']],
[[id: 'batch3', date: '25.11.14'], ['file3A.txt', 'file3B.txt']]]
Then the output channel contents coming out like this:
ch_input = [[[id: 'batch1', date: '25.10.01'], 'batch1.txt'],
[[id: 'batch2', date: '25.10.26'], 'batch2.txt'],
[[id: 'batch3', date: '25.11.14'], 'batch3.txt']]
As mentioned earlier, the tuple val(meta), path(files_in) input setup is a standard pattern used across all nf-core modules.
Hopefully you can start to see how useful this can be. Not only does it allow you to name outputs based on metadata, but you can also do things like use it to apply different parameter values, and in combination with specific operators, you can even group, sort or filter out data as it flows through the pipeline.
Learn more about metadata
For a comprehensive introduction to working with metadata in Nextflow workflows, including how to read metadata from samplesheets and use it to customize processing, see the Metadata in workflows side quest.
2.3. Summarize changes to be made¶
Based on what we've reviewed, these are the major changes we need to make to our pipeline to utilize the cat/cat module:
- Create a metamap containing the batch name;
- Package the metamap into a tuple with the set of input files to concatenate (coming out of
convertToUpper); - Switch the call from
collectGreetings()toCAT_CAT; - Extract the output file from the tuple produced by the
CAT_CATprocess before passing it tocowpy.
That should do the trick! Now that we've got a plan, we're ready to dive in.
Takeaway¶
You know how to assess the input and output interface of a new module to identify its requirements, and you've learned how metamaps are used by nf-core pipelines to keep metadata closely associated with the data as it flows through a pipeline.
What's next?¶
Integrate the new module into a workflow.
3. Integrate CAT_CAT into the hello.nf workflow¶
Now that you know everything about metamaps (or enough for the purposes of this course, anyway), it's time to actually implement the changes we outlined above.
For the sake of clarity, we'll break this down and cover each step separately.
Note
All the changes shown below are made to the workflow logic in the main block in the core-hello/workflows/hello.nf workflow file.
3.1. Create a metadata map¶
First, we need to create a metadata map for CAT_CAT, keeping in mind that nf-core modules require the metamap to at least an id field.
Since we don't need any other metadata, we can keep it simple and use something like this:
Except we don't want to hardcode the id value; we want to use the value of the params.batch parameter.
So the code becomes:
Yes, it is literally that simple to create a basic metamap.
Let's add these lines after the convertToUpper call, removing the collectGreetings call:
| core-hello/workflows/hello.nf | |
|---|---|
This creates a simple metadata map where the id is set to our batch name (which will be test when using the test profile).
3.2. Create a channel with metadata tuples¶
Next, transform the channel of files into a channel of tuples containing metadata and files:
| core-hello/workflows/hello.nf | |
|---|---|
The line we've added achieves two things:
.collect()gathers all files from theconvertToUpperoutput into a single list.map { files -> tuple(cat_meta, files) }creates a tuple of[metadata, files]in the formatCAT_CATexpects
That is all we need to do to set up the input tuple for CAT_CAT.
3.3. Call the CAT_CAT module¶
Now call CAT_CAT on the newly created channel:
This completes the trickiest part of this substitution, but we're not quite done yet: we still need to update how we pass the concatenated output to the cowpy process.
3.4. Extract the output file from the tuple for cowpy¶
Previously, the collectGreetings process just produced a file that we could pass to cowpy directly.
However, the CAT_CAT process produces a tuple that includes the metamap in addition to the output file.
Since cowpy doesn't accept metadata tuples yet (we'll fix this in the next part of the course), we need to extract the output file from the tuple produced by CAT_CAT before handing it to cowpy:
The .map{ meta, file -> file } operation extracts the file from the [metadata, file] tuple produced by CAT_CAT into a new channel, ch_for_cowpy.
Then it's just a matter of passing ch_for_cowpy to cowpy instead of collectGreetings.out.outfile in that last line.
Note
In the next part of the course, we'll update cowpy to work with metadata tuples directly, so this extraction step will no longer be necessary.
3.5. Test the workflow¶
Let's test that the workflow works with the newly integrated cat/cat module:
This should run reasonably quickly.
Output
N E X T F L O W ~ version 25.04.3
Launching `./main.nf` [evil_pike] DSL2 - revision: b9e9b3b8de
Input/output options
input : /workspaces/training/hello-nf-core/core-hello/assets/greetings.csv
outdir : core-hello-results
Institutional config options
config_profile_name : Test profile
config_profile_description: Minimal test dataset to check pipeline function
Generic options
validate_params : false
trace_report_suffix : 2025-10-30_18-50-58
Core Nextflow options
runName : evil_pike
containerEngine : docker
launchDir : /workspaces/training/hello-nf-core/core-hello
workDir : /workspaces/training/hello-nf-core/core-hello/work
projectDir : /workspaces/training/hello-nf-core/core-hello
userName : root
profile : test,docker
configFiles : /workspaces/training/hello-nf-core/core-hello/nextflow.config
!! Only displaying parameters that differ from the pipeline defaults !!
------------------------------------------------------
executor > local (8)
[b3/f005fd] CORE_HELLO:HELLO:sayHello (3) [100%] 3 of 3 ✔
[08/f923d0] CORE_HELLO:HELLO:convertToUpper (3) [100%] 3 of 3 ✔
[34/3729a9] CORE_HELLO:HELLO:CAT_CAT (test) [100%] 1 of 1 ✔
[24/df918a] CORE_HELLO:HELLO:cowpy [100%] 1 of 1 ✔
-[core/hello] Pipeline completed successfully-
Notice that CAT_CAT now appears in the process execution list instead of collectGreetings.
And that's it! We're now using a robust community-curated module instead of custom prototype-grade code for that step in the pipeline.
Takeaway¶
You now know how to:
- Find and install nf-core modules
- Assess the requirements of an nf-core module
- Create a simple metadata map for use with an nf-core module
- Integrate an nf-core module into your workflow
What's next?¶
Learn to adapt your local modules to follow nf-core conventions. We'll also show you how to create new nf-core modules from a template using the nf-core tooling.